![]()

|
|
 |
 My
research interests are focused on the genetic and molecular analysis
of chromosome and chromatin structure in Drosophila
melanogaster. Through examining and understanding mutations
associated with gene position effects, my work will help elucidate
the role that chromosome and chromatin structure plays in regulating
gene expression. I am currently concentrating on the cubitus
interruptus (ci) locus on Drosophila chromosome 4 and its
unusual position effects. I am also involved in the large scale
physical mapping of chromosome 4 .
My
research interests are focused on the genetic and molecular analysis
of chromosome and chromatin structure in Drosophila
melanogaster. Through examining and understanding mutations
associated with gene position effects, my work will help elucidate
the role that chromosome and chromatin structure plays in regulating
gene expression. I am currently concentrating on the cubitus
interruptus (ci) locus on Drosophila chromosome 4 and its
unusual position effects. I am also involved in the large scale
physical mapping of chromosome 4 .
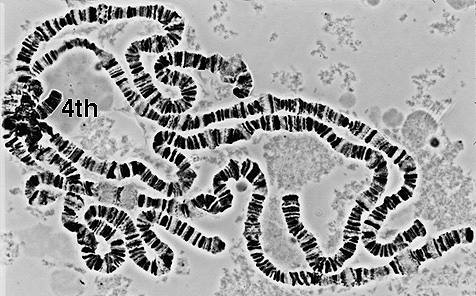 The
model organism Drosophila melanogaster has two sex chromosomes
and 3 autosomes.The smallest chromosome is chromosome 4, ~5 Mbp in
length (Locke and McDermid, 1993), which appears as a "dot"
chromosome in metaphase spreads. Chromosome 4 has two major regions.
The centromeric domain is a-heterochromatic and consists primarily of
about ~3-4 Mbp of short, satellite repeats. This region forms part of
the highly condensed chromocenter seen in polytene chromosome
spreads. The remaining ~1.2 Mbp constitutes cytogenetic regions 101E
to 102F on salivary gland chromosomes (the banded region). Of the ~80
genes expected on chromosome 4, only 15-20 of the genes have been
mapped and so far all to this domain.
The
model organism Drosophila melanogaster has two sex chromosomes
and 3 autosomes.The smallest chromosome is chromosome 4, ~5 Mbp in
length (Locke and McDermid, 1993), which appears as a "dot"
chromosome in metaphase spreads. Chromosome 4 has two major regions.
The centromeric domain is a-heterochromatic and consists primarily of
about ~3-4 Mbp of short, satellite repeats. This region forms part of
the highly condensed chromocenter seen in polytene chromosome
spreads. The remaining ~1.2 Mbp constitutes cytogenetic regions 101E
to 102F on salivary gland chromosomes (the banded region). Of the ~80
genes expected on chromosome 4, only 15-20 of the genes have been
mapped and so far all to this domain.
The fourth is an atypical Drosophila chromosome in several respects, all of which relate to the long-standing presumption that this chromosome is, in some ways, heterochromatic in nature. First, the banded region of chromosome 4 often shows a diffuse and poorly defined appearance similar to regions of b -heterochromatin. Second, a similarity to b-heterochromatin is shown in that the chromosomal protein HP1, thought to be an important constituent of heterochromatin, binds to several sites along the chromosome. Third, repetitive DNA sequences normally confined to b -heterochromatin are distributed throughout the banded region of 4. Fourth, further similarities to b -heterochromatin are indicated by the behavior of P element transgenes inserted into this region since they frequently show variegated expression of a white+ marker gene, a characteristic of insertion sites near heterochromatic boundaries.Fifth, chromosome 4 lack crossing over during meiosis.
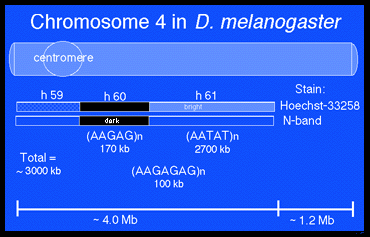
The loci on chromosome 4 have been shunned in the Drosophila literature. Most genetic screens have been designed to recover mutants on only the X, 2, or 3 and ignored chromosome 4 loci. This evasive action was probably due to its small size and the last feature above, lack of crossing over. Without crossing over was perceived as hard, if not impossible, to characterize chromosome 4 genes. Nevertheless, Hochman in 1976 identified ~ loci and roughly mapped them using the few deletions available at the time.
 Chromosome
4, which constitutes cytogenetic regions 101E to 102F on salivary
gland chromosomes, is the region we have decided to map. Beginning
with many entry points and many chromosomal walks we have recovered
overlapping cosmid and BAC clones that together constitute a contig
that spans the length of the "banded region" defined by cytogenetic
bands 101E to 102F. Chromosome walking has been hindered by the
abundance of moderately repeated sequences dispersed along the
chromosome. The clones have been correlated to the cytogenetic map by
in situ hybridization to polytene chromosomes. The locations of
previously cloned genes have also been mapped on the contig. A
minimal tiling set of clones from the contig will provide templates
for sequencing this chromosome as part of the Drosophila Genome
Project.
Chromosome
4, which constitutes cytogenetic regions 101E to 102F on salivary
gland chromosomes, is the region we have decided to map. Beginning
with many entry points and many chromosomal walks we have recovered
overlapping cosmid and BAC clones that together constitute a contig
that spans the length of the "banded region" defined by cytogenetic
bands 101E to 102F. Chromosome walking has been hindered by the
abundance of moderately repeated sequences dispersed along the
chromosome. The clones have been correlated to the cytogenetic map by
in situ hybridization to polytene chromosomes. The locations of
previously cloned genes have also been mapped on the contig. A
minimal tiling set of clones from the contig will provide templates
for sequencing this chromosome as part of the Drosophila Genome
Project.
Under construction.

|
|
 |
Papers: Submitted, in press, or published. (Jan'00)
McFadyen, D. A., Locke, J.
High resolution FISH mapping of the rat a2u-globulin multigene
family.
Mammalian Genome (at the Press) 1999.
J. Locke, L.T. Howard, N.
Aippersbach, L. Podemski and R.B. Hodgetts (1999)
The characterization of DINE-1, a short, interspersed repetitive
element present on chromosome 4 and in the centric heterochromatin of
Drosophila melanogaster .
Chromosoma 108:356-366.
Locke, J., L. Podemski , K. Roy, D. Pilgrim and R. Hodgetts
(1999)
DNA sequence analysis of two cosmid clones from chromosome 4 of
Drosophila melanogaster predicts two new genes amid an unusual
arrangement of repeated sequences.
Genome Research 9;137-149
McFadyen, D. A. W. Addison, and J. Locke (1999)
Genomic Organization of the Rat a2u-Globulin
Gene Cluster.
Mammalian Genome 10:463-470.
Wang K.S., McFadyen D.A., Locke J, Hodgetts R.B. (1997)
Three subsets of genes whose tissue specific expression is sex and
age-dependent can be identified within the rat alpha 2u-globulin
family.
Dev. Genet 21:234-244
 Locke, J. and S. Hanna. (1996)
Locke, J. and S. Hanna. (1996)
engrailed gene dosage determines whether certain recessive
cubitus interruptus alleles exhibit dominance of the adult
wing phenotype in Drosophila.
Dev. Genetics 19:340-349.
Locke, J., Rairdan, G., McDermid, H., Nash, D., Pilgrim,
D., Bell, J. Roy, K., and R. Hodgetts. (1996)
Cross-screening: a new method to assemble clones rapidly and
unambiguously into contigs.
Genome Research 6:155-165.
Schwartz, C., Locke, J., Nishida, C. and T. Kornberg.
(1995)
Analysis of cubitus interruptus regulation in
Drosophila embryos and imaginal disks.
Development 121;1625-1635.
Locke, J. and K.D. Tartof (1994).
Molecular analysis of cubitus interruptus (ci) mutations
suggests an explanation for the unusual ci-position
effect.
Molecular and General Genetics 243: 234-243.
Locke, J. and McDermid, H. E. (1993)
Analysis of Drosophila Chromosome 4 using pulsed field gel
electrophoresis.
Chromosoma 102:718-723.
Locke, J. (1993)
Examination of DNA sequences undergoing chromatin changes at a
variegating breakpoint in Drosophila melanogaster.
Genetica 92: 33-41.
Locke, J., S. Hanna and D. Kong. (1993).
Evidence for mobilization of hobo transposons in a P-element
mutagenesis screen.
Genome 36:1138-1147.
Tartof, K.D. Bishop, C., Jones, M., Hobbs, C.A., and Locke,
J. (1989).
Towards an understanding of position effect variegation.
Dev. Genetics 10; 162-176.
Locke, J., Kotarski, M.A., and Tartof, K. D. (1988).
Dosage dependent modifiers of position effect variegation in
Drosophila and a mass action model that explains their effect.
Genetics 120;181-198.
Copyright © 1999 John Locke